Introduction
Estimating the the models described in Explaining Recruitment to Violent Extremism: A Bayesian Case-Control Approach (ERVE) requires just two steps:
- Prepare the data sources in list format using
data.prep() - Estimate the model using
fit()
This explainer vignette walks you through the first of these two steps.
Preparing the dataset for the estimation procedure requires two sources of data: 1) survey data with labels for “cases” and “controls” (as well as any relevant covariates) and; a shapefile for the geographical region under investigation.
Setup
First install the package from source if not already installed.
devtools::install_github("extremerpkg/extremeR")We then load the package into memory as follows.
We can then load the data we need into memory using the data bundled in this package as follows.
We can see that our case-control survey data,
survey_egypt, and our denominator data,
denom_mena are “data.table” “data.frame” objects.
Our Egypt shapefile data is here stored as an “sf” “data.frame” object.
class(survey_egypt)## [1] "data.table" "data.frame"
class(shape_egypt)## [1] "sf" "data.frame"
class(denom_mena)## [1] "data.table" "data.frame"Note that here we are using data bundled with the extremeR package. If we had a series of shapefiles of the type provided e.g. here, we could generate the “sf” object from these files as follows.
shapefile_path <- "data/EGY_adm2.shp"
shape_egypt <- sf::st_read(shapefile_path)Inspect data
Now that we have each data source that we need: i.e., the case-control data and the shapefile, we are ready to inspect the data. We will begin with the case-control data.
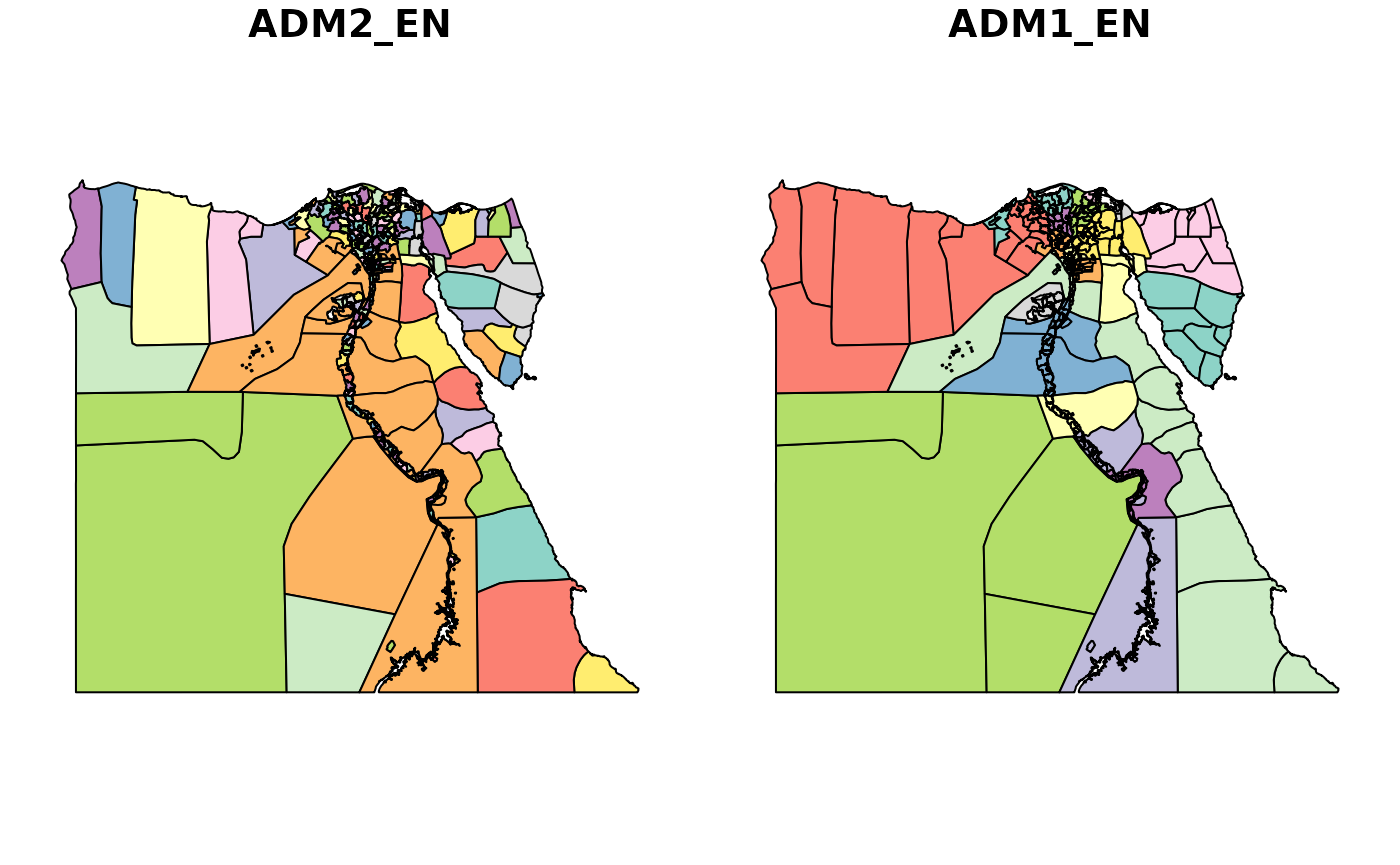
The shapefile data is plotted below. The relevant geographic identifiers we’ll be using in the shapefile are “ADM1_EN” for the large area and “ADM2_EN” for the small area.
We’ll then be merging with the “ADM2_PCODE” identifier, as this is the common identifier between our shapefile and case-control data.
Finally, we need to input information on the known prevalence of the outcome in the population of interest. For this, we need to know the relevant population denominator. In the ERVE example, this is the adult male Sunni population.
These data are also bundled into the package, and we can look at them below.To extract the relevant denominator for Egypt we can do as follows.
denom_egypt <- denom_mena$male_18_sunni[which(denom_mena$country=="Egypt")]Preparing the data
We are now in a position to bundle these data sources into a list
object suitable for the fit() function to estimate one of
the models described in ERVE.
The data.prep() function has a series of parameters that
we tabulate and describe below.
| Argument | Description |
|---|---|
| shape | sf object, shapefile data |
| survey | data.table data.frame, case-control data including common geographic ID |
| shape_large.area_id_name | string, large area name identifiers in the shapefile |
| shape_large.area_id_num | integer, large area identifiers in the shapefile |
| shape_small.area_id_name | string, small area name identifiers in the shapefile |
| shape_small.area_id_num | integer, small area identifiers in the shapefile |
| survey_small.area_id_name | string, small area name identifiers in the survey |
| survey_small.area_id_num | integer, small area identifiers in the survey |
| drop.incomplete.records | logical, should the function return complete data?
Defaults to TRUE
|
| colnames_X | character vector, covariates definining the design matrix X. Must be numeric. |
| interactions_list | list, each element is a string of the form “a*b” where a and be are the names of two variables in colnames_X |
| scale_X | string, takes values “1sd” or “2sd” |
| colname_y | string, variable name for the outcome variable. Must be numeric. |
| contamination | logical, should this offset account for
contamination? Defaults to TRUE
|
| pi | numeric, scalar defining the prevalence of the outcome in the population of interest |
| large_area_shape | logical, should the function return a large-area
shapefile? Defaults to TRUE
|
# prep data
data.list =
data.prep(
shape = shape_egypt,
survey = survey_egypt,
shape_large.area_id_name = "ADM1_EN",
shape_large.area_id_num = NA,
shape_small.area_id_name = "ADM2_EN",
shape_small.area_id_num = "ADM2_PCODE",
survey_small.area_id_name = NA,
survey_small.area_id_num = "adm2_pcode",
drop.incomplete.records = T,
colnames_X = c(
"coledu",
"age",
"married",
"student",
"lowstat",
"population_density",
"total_population_2006",
"christian_2006_pct",
"university_2006_pct",
"agriculture_2006_pct",
"mursi_vote_2012_pct",
"sqrt_killed_at_rabaa",
"unemployment_2013q4_pct",
"sqrt_protest_post_Mubarak"
),
interactions_list = list(age2 = "age*age", coledu_lowstat = "coledu*lowstat"),
scale_X = "1sd",
colname_y = "case",
contamination = T,
pi = 1000 / denom_egypt,
large_area_shape = T
)The data.prep() function takes our shapefile and
case-control data and bundles it into a list.
We can look at the structure of these data below. We see that the list contains our survey data, shapefiles, generated neighbourhood matrices, outcome observations etc.
summary(data.list)## Length Class Mode
## survey_complete 20 data.table list
## n 1 -none- numeric
## y 617 -none- numeric
## pi 1 -none- numeric
## n1 1 -none- numeric
## n0 1 -none- numeric
## offset 1 -none- numeric
## theta_1 1 -none- numeric
## theta_0 1 -none- numeric
## X 17 data.table list
## p 1 -none- numeric
## mu_X 16 -none- numeric
## sd_X 16 -none- numeric
## scale_X 1 -none- character
## small_area_id 617 -none- numeric
## small_area_names 365 -none- character
## large_area_id 617 -none- numeric
## large_area_names 27 -none- character
## N_large_area 1 -none- numeric
## nb_object 365 nb list
## N_small_area 1 -none- numeric
## node1_small_area 824 -none- numeric
## node2_small_area 824 -none- numeric
## N_small_area_edges 1 -none- numeric
## scaling_factor 1 -none- numeric
## nb.matrix 133225 -none- numeric
## shape_small 20 sf list
## shape_large 2 sf listAnd we can save the formatted data for later use as follows.
save(data.list, "data/data.list.rda")We can then check that the neighbourhood matrix is properly connected
using the testconnected() function as below.
# check that the nb object is fully connected
testconnected(data.list$nb_object)## [1] "Success! The Graph is Fully Connected"