Introduction
Estimating the the models described in Explaining Recruitment to Violent Extremism: A Bayesian Case-Control Approach (ERVE) requires just two steps:
- Prepare the data sources in list format using
data.prep() - Estimate the model using
fit()
This explainer vignette walks you through the second of these two steps.
Setup
First install the package from source if not already installed.
devtools::install_github("extremerpkg/extremeR")We then load the package into memory as follows. We will also be
using several functions from the rstan package.
Estimate model
And we are then ready to estimate the ERVE model for the Egyptian case-control data.
The fit() function has a series of parameters that we
tabulate and describe below.
| Argument | Description |
|---|---|
| data | list, survey and shapefile combined list object
created with the data.prep() function |
| show_code | logical, should the stan code print on the screen at
the end of the run? Defaults to TRUE
|
| contamination | logical, should the model include the Rota et
al. (2013) style contamination layer? Defaults to TRUE
|
| offset | logical, should the model include a King and Zeng
(2001) style offset ? if contamination is also specified, this will be a
contaminated-offset. Defaults to TRUE
|
| beta_prior | string, what prior should the regression coefficients have? Choice between: “normal” and “cauchy” |
| small_area_prior | string, what should be the small-area effects type?
Choice between: “fixed,” “random,” “ICAR,” and “BYM2.” Specify
NA for no area effects. Defaults to “fixed” |
| large_area_prior | string, what should be the large-area effects type?
Choice between: “fixed” and “random.” Specify NA for no
area effects. Defaults to NA. |
| intercept_scale | integer, scale of intercept; arguments taken from
rstan, see here for further
information |
| iter | integer, number of draws; arguments taken from
rstan, see here for further
information |
| warmup | integer, number of warmup draws; arguments taken from
rstan, see here for further
information |
| thin | integer, number to thin draws; arguments taken from
rstan, see here for further
information |
| cores | integer, number of cores to use; arguments taken from
rstan, see here for further
information |
| chains | integer, number of chains; arguments taken from
rstan, see here for further
information |
| control | list, tree depth and adapt delta; arguments taken
from rstan, see here for further
information |
| verbose | logical, print full stan output? Defaults to
TRUE
|
fit_object = fit(
data = data.list,
show_code = T,
contamination = T,
offset = T,
beta_prior = "cauchy",
small_area_prior = "BYM2",
intercept_scale = 10,
large_area_prior = "random",
iter = 25000,
warmup = 22500,
thin = 4,
cores = 4,
chains = 4,
control = list(max_treedepth = 25, adapt_delta = 0.99),
verbose = T
)Check MCMC convergence
We can then check the summary convergence statistics by using the
monitor() function in rstan.
And we could inspect the output for, e.g., the \(\hat{R}\) values as follows.
rhats <- mon_egypt$Rhat
plot(rhats)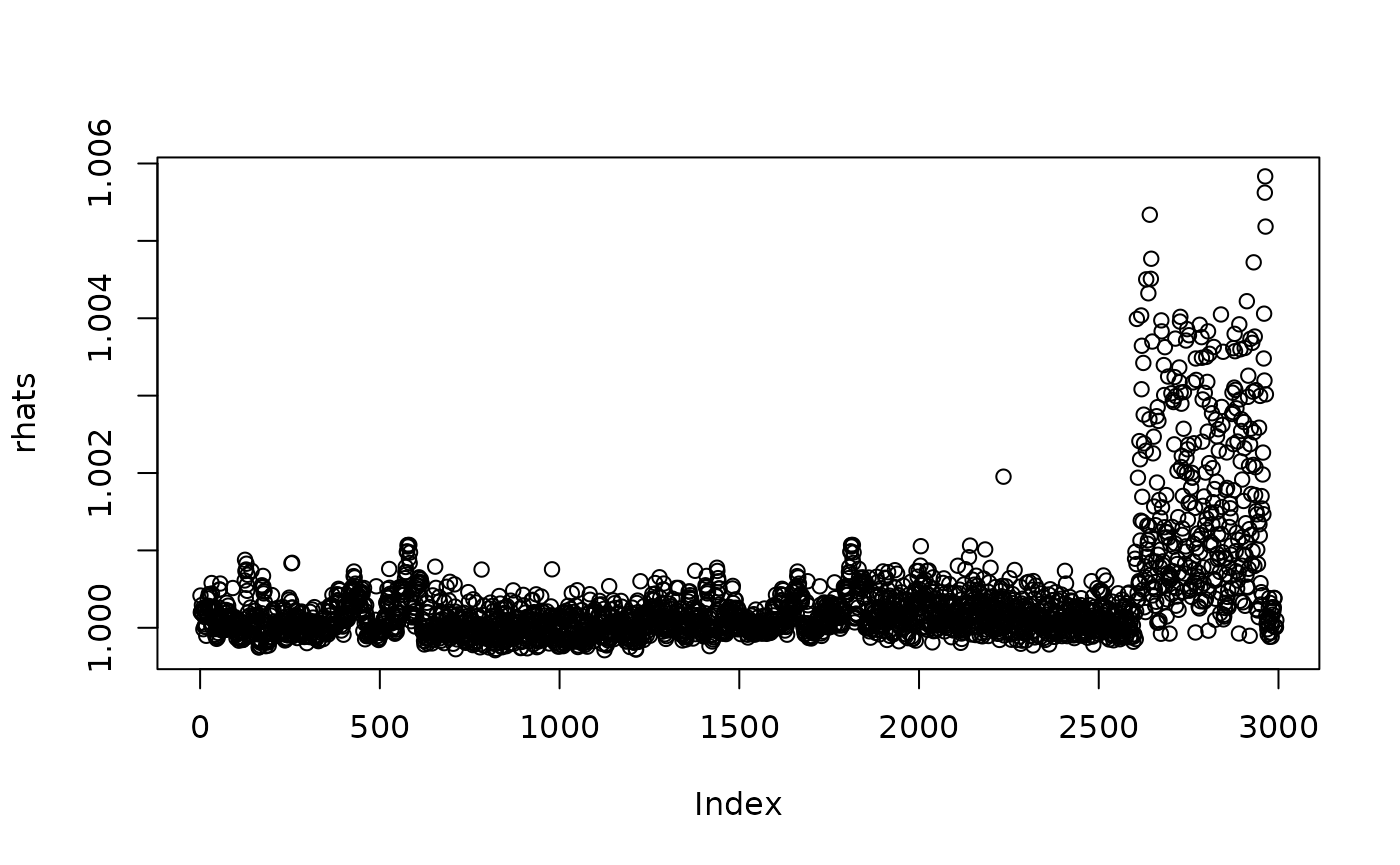
Inspect output
And we are able to access the estimated parameters of the “stanfit”
object by using the extract() function in
rstan.
betas_egypt <- rstan::extract(fit_object ,
pars = c('beta'),
permuted = TRUE,
inc_warmup = FALSE,
include = TRUE)Or printing from the fit object directly.
## Inference for Stan model: anon_model.
## 4 chains, each with iter=25000; warmup=22500; thin=4;
## post-warmup draws per chain=625, total post-warmup draws=2500.
##
## mean se_mean sd 10% 50% 90% n_eff Rhat
## beta[1] -13.26 0.02 0.88 -14.41 -13.19 -12.20 2265 1
## beta[2] 0.56 0.01 0.27 0.22 0.55 0.91 2683 1
## beta[3] -0.38 0.02 1.05 -1.65 -0.39 0.84 2513 1
## beta[4] 0.05 0.01 0.31 -0.35 0.06 0.45 2361 1
## beta[5] 0.18 0.00 0.23 -0.11 0.18 0.47 2462 1
## beta[6] -0.85 0.01 0.47 -1.46 -0.81 -0.30 2470 1
## beta[7] 1.36 0.01 0.73 0.49 1.27 2.31 2472 1
## beta[8] -0.15 0.01 0.57 -0.87 -0.15 0.57 2004 1
## beta[9] -0.29 0.01 0.61 -1.05 -0.27 0.43 1698 1
## beta[10] 0.29 0.01 0.60 -0.44 0.27 1.04 2193 1
## beta[11] -0.84 0.02 0.86 -1.96 -0.74 0.16 2318 1
## beta[12] -0.09 0.02 0.72 -0.99 -0.06 0.77 1873 1
## beta[13] 1.59 0.02 0.73 0.68 1.57 2.54 1674 1
## beta[14] -0.26 0.01 0.64 -1.09 -0.23 0.49 2173 1
## beta[15] -0.39 0.01 0.60 -1.21 -0.34 0.33 2208 1
## beta[16] -1.15 0.03 1.26 -2.75 -0.99 0.24 2457 1
## beta[17] 0.48 0.00 0.26 0.17 0.47 0.82 2674 1
##
## Samples were drawn using NUTS(diag_e) at Tue Mar 1 23:48:57 2022.
## For each parameter, n_eff is a crude measure of effective sample size,
## and Rhat is the potential scale reduction factor on split chains (at
## convergence, Rhat=1).